Welcome to the Maguire Lab
We are a research group jointly based in Dalhousie’s Faculty of Computer Science and the Faculty of Medicine’s Department of Community Health & Epidemiology.
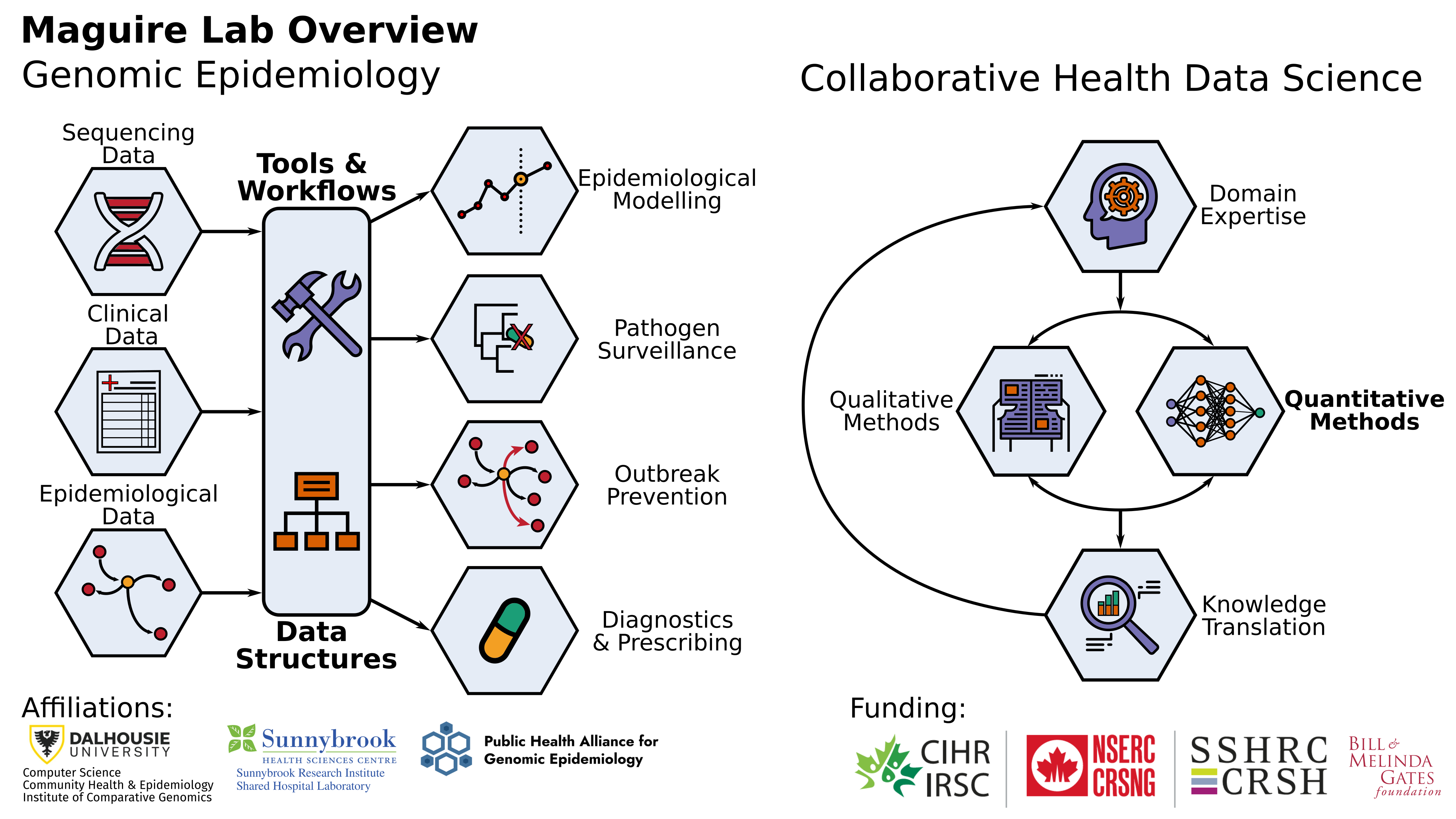
Our aim is to develop and collaboratively apply data-driven methods to try and mitigate health and social crises. This is focused on two main areas: genomic epidemiology of infectious diseases and inter-disciplinary collaborations with domain experts
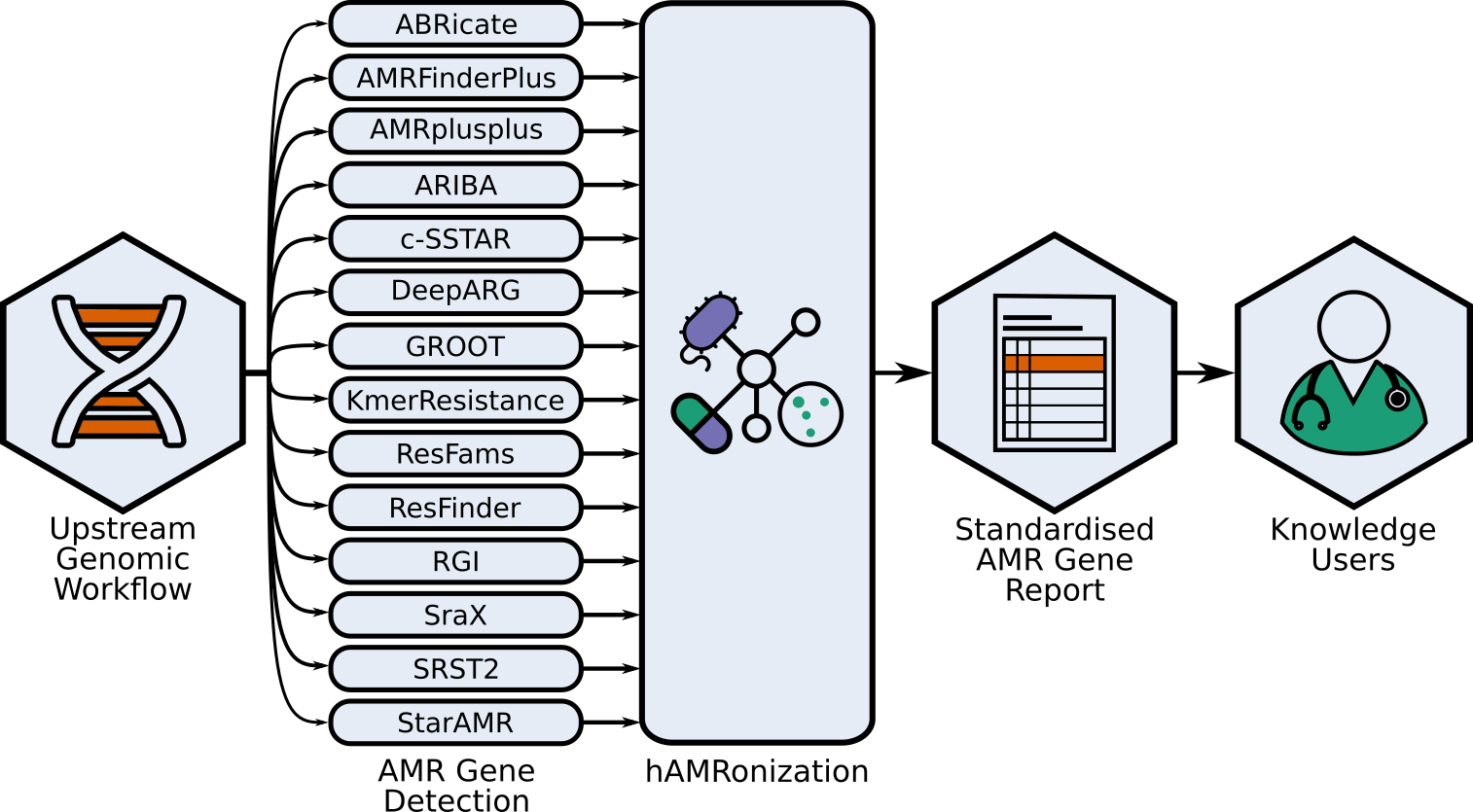
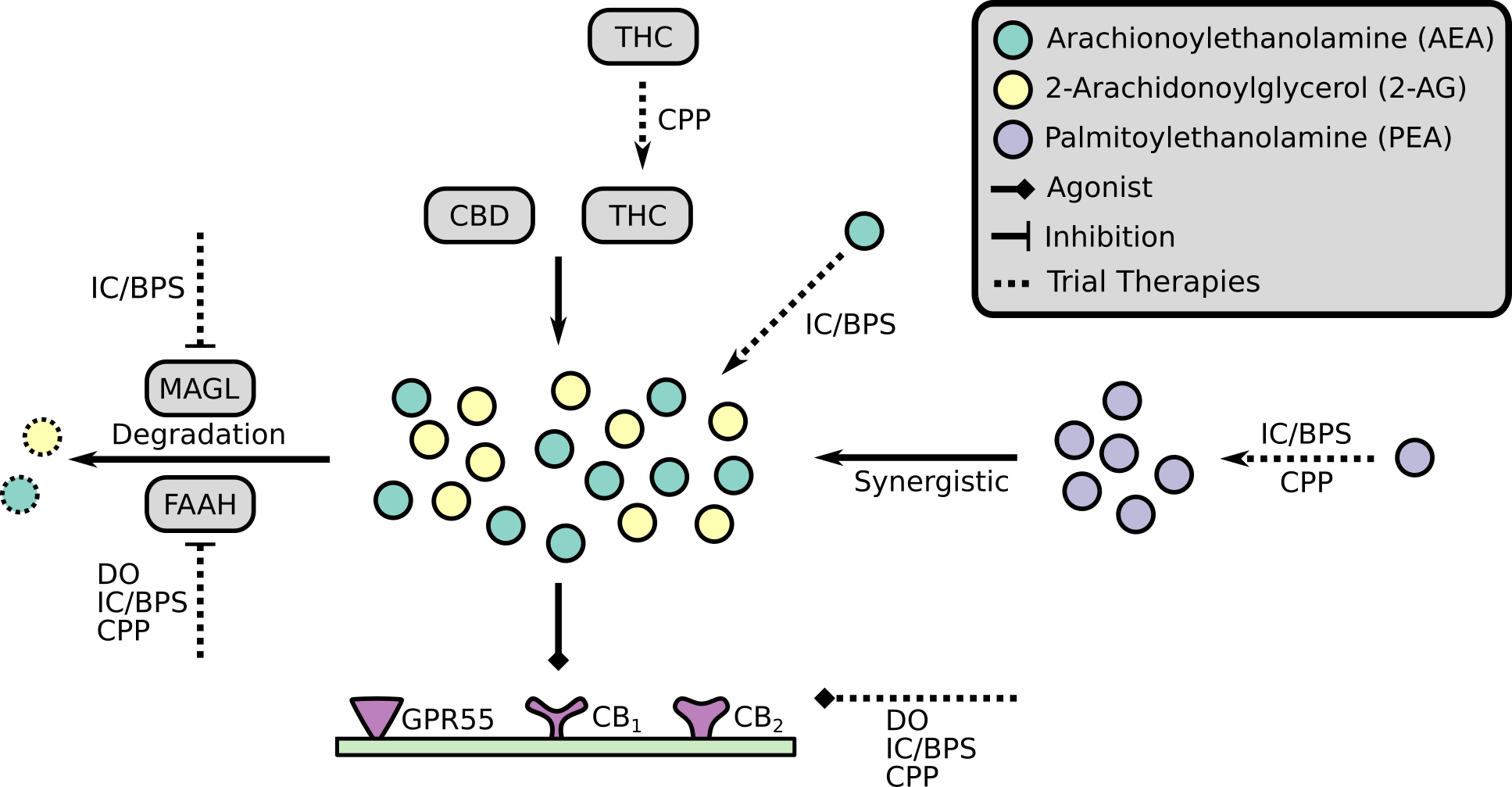
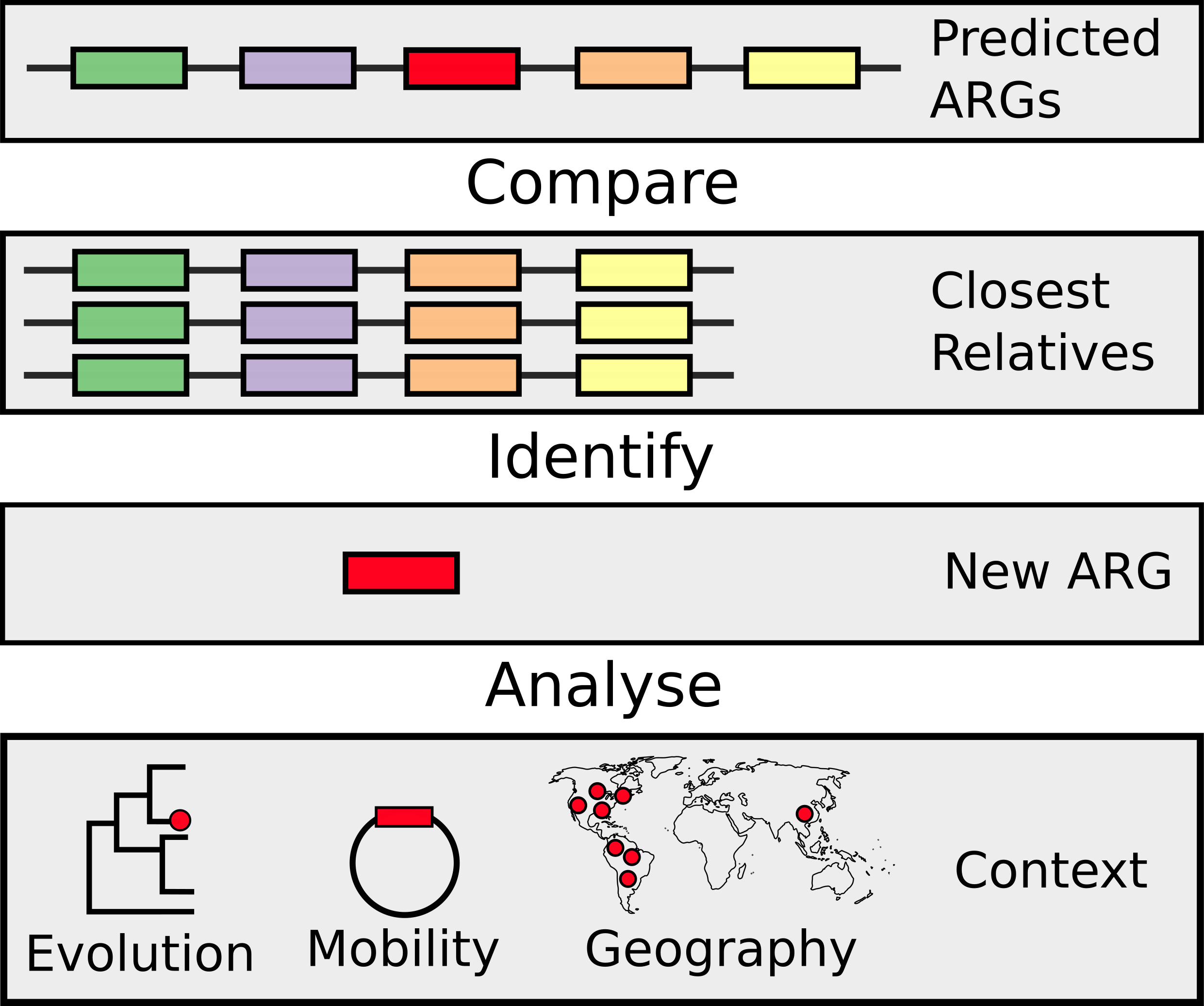
Specifically, this former work involves developing and applying novel microbial bioinformatics and machine learning approaches to better understand the diagnosis, evolution, and dynamics of infectious diseases. We largely work on problems related to antimicrobial resistance (AMR) and, in the last couple of years, the COVID-19 pandemic with national and international consortia of clinicians and public health experts. Whereas, our broader collaborative data science works includes work exploring online radicalisation with sociologists, patient preference at refugee clinics, and autism-related language-use.
For more details about specific projects, collaborators, and funding sources see Research).
We are located in Dalhousie University and have strong ties to the Shared Hospital Lab located at Sunnybrook Health Sciences Centre, the CARD, the Canadian Food Inspection Agency, and the Public Health Agency of Canada. This also includes national and international public health consortia such as IRIDA, CanCOGeN, and PHA4GE.
Joining the Lab
We are looking for new enthusiastic and creative PhD students to join the team!
Funding
We are grateful for funding from Dalhousie University, CIHR, NSERC, CANMOD, Genome Canada, SSHRC, and (via PHA4GE) the BMGF.

News
25 December 2025David Mahoney’s first PhD chapter benchmarking different methods for querying assembly graphs was released as a pre-print on BioRxiv
Yann Guerin’s first paper was published in Journal of Ob/Gyn Canada. We also published a paper on episiotomy misinformation on TikTok with the same collaborators
A pair of papers from the clinical lab were published recently on beta-lactamase genotype-phenotype correlation in Pseudomonas, nirmatrelvir resistance, and viral metagenomic surveillance
Finlay Maguire was elected as a member of the Royal Society of Canada’s College of New Scholars, Artists and Scientists
In work led by Dr. Stairs, our paper on the primary management for stress urinary incontinence by socioeconomic status was published in JOGC
2 preprints were released: the PHA4GE wastewater surveillance data specification (led by Jillian Paull and Emma Griffiths), and Lauran Crawshaw and Jeff Bowman’s WildEPI work searching for a B.1.641 reservoir in deer
Finlay Maguire gave an invited talk as ASM Microbe in Los Angeles on the interpretation and contextualisation of AMR genotypic results
David Mahoney was awarded a Canadian Bioinformatics Hub Training Award to support attendance of a relevant workshop or conference
Two SARS-CoV-2 papers from the clinical lab were recently published: Natalie Deschenes paper in the Journal of Infectious Disease on Nirmatrelvir resistance mutations and Kuganya Nirmalarajah’s paper in BMC Infectious Diseases on severe infection predictive factors